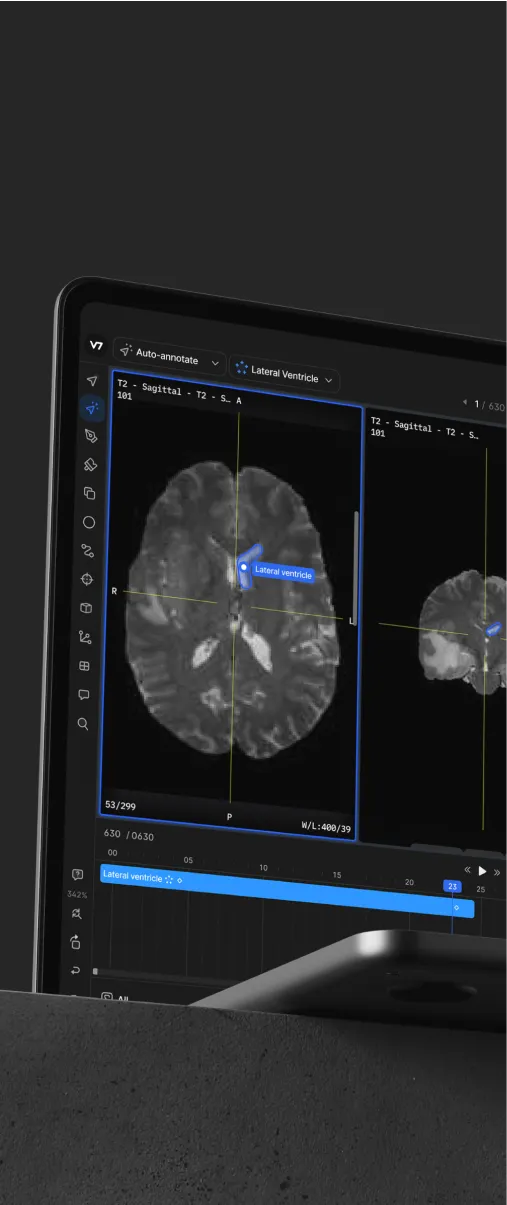
Playbooks
9 min read
—
Jan 24, 2023
Are you a pathologist looking for ways to improve your annotation of tissue samples? This article provides best practices and tips to help you speed up the process. Learn how to label and organize datasets, use auto-annotation tools, and benefit from existing datasets for digital pathology tasks.

Casimir Rajnerowicz
Content Creator
Machine learning is changing the way healthcare professionals diagnose and treat diseases. And if there is a medical field in which this is particularly evident, it is digital pathology.
Computer vision models can now detect diseases such as diabetic retinopathy or breast cancer with remarkable accuracy. AI is able to recognize subtle differences between normal and abnormal tissue. It can go through a sample and check every single cell for signs of cancer in a fraction of the time that a pathologist would need. AI models can also identify patterns in medical images that are too complex for the human eye to notice.
However, in order for these models to be accurate and reliable, they must be trained on large datasets of annotated digital pathology samples.
The process usually looks something like this:
Step 1: Labeling your data with semi-automatic segmentation tools

Step 2: Training an AI classification and instance segmentation model

Step 3: Using the model for automated analysis of your new data

In the example above, we trained a model to segment red blood cells and leukocytes. We can use it to analyze histological slides and get RBC-to-leukocyte ratio in seconds. With some additional tweaks, the model can detect blood cell formation patterns.
While this sort of task is surprisingly easy to complete with digital pathology annotation software, there are many things that can go wrong.
This article outlines the best practices and challenges associated with annotating digital pathology images for AI projects.
Let’s start with some potential obstacles that you should be aware of as a pathologist.
Challenges of labeling digital pathology data
Pathology images can be very complex and may contain a wide range of different structures and patterns. This can make it difficult to accurately label and annotate the samples, as it requires a deep understanding of the underlying biology and pathology.
Common challenges of annotating tissue samples and training AI models for digital pathology include:
Lack of clear instruction or label management. There is often a lack of standardization in the way that datasets, labels, and annotation classes are defined and organized. The importance of dataset management is usually discovered late in the project. And it usually leads to costly delays.
Manual slide annotation is time-consuming. Outlining regions of interest point by point is a tedious and labor-intensive process. Most auto-annotation software available on the market still struggles with recognizing cells and other features in microscopy images.
Large size of the datasets and unique file formats. Digital pathology images, such as WSI files can be very large. A single image that is several gigabytes is not uncommon. This can make it difficult to store and manage large datasets, especially if you are working with a large number of slides.
Limited availability of expert annotators. To understand the data and provide meaningful labels domain expertise is sometimes an absolute must. Expert annotators, such as pathologists, are typically in high demand and may not be readily available to annotate large datasets.
The review process frequently needs multiple stages. Different pathologists may label the same image differently. This can lead to inconsistencies in the dataset and affect the performance of the AI model. But even when multiple experts are involved in the annotation process, there may still be disagreements about the labels or annotations for a given image. Setting up a data annotation workflow with specific roles based on seniority is usually necessary.
It is hard to test AI models with small datasets. The process from annotation to model testing can be quite lengthy. And it can be difficult to validate the effectiveness of your project without a significant amount of effort. To make things harder, most AI training platforms are not designed to work with small datasets.
Thankfully, you can address all of these problems with careful planning and the right tools.
Here are some best practices for working with digital pathology images:
1. Use labels and annotation classes that are right for the task
Determine what you are going to analyze at the very beginning of the project. To detect and measure the size of a granule, you might want to decide on polygons or bounding box annotations. But if we need to track the movement of bacteria, keypoint skeletons may produce more accurate models.
Some annotation classes are suitable for measuring growth or trajectory while others are better suited for tasks such as object classification.

It is also important to establish some rules for labeling similar structures. For instance, you can create one class for a specific cell type but use additional attributes such as “normal”, “abnormal”, “malignant”, etc. Alternatively, in some scenarios, it might be better to disregard class attributes and create“normal cell” and “cancer cell” as two separate classes. It is up to you, but make sure to stick to one method.
In V7 you can choose your annotation classes, labels, and attributes while setting up a new dataset annotation project. Then you can block other users from creating new label types on their own.

This can help you control the annotation process and ensure that your data is consistent and valid. Just remember to write an instruction for your annotators and explain which objects need labeling with and with labels.
Read more: Best Practices for Writing Annotation Instructions
2. Add review and consensus stages to your annotation workflows
Pathologists may have different interpretations of the same slide. It is important to resolve any discrepancies to ensure that the dataset is of high quality. Consensus stages and reviews can help to identify and address any disagreements about the labels for a given tissue sample. They can also help to identify any biases.

For instance, you can decide what level of annotation overlap (for instance, based on Intersection Over Union) is acceptable. The same slide is then labeled by multiple annotators who cannot see each other's annotations. The annotations are then compared to determine the level of agreement between the annotators. If the level of agreement is below the acceptable threshold, the annotations are reviewed by a senior pathologist or a panel of experts.

Consensus stages can also be helpful for testing the performance of AI models. During later iterations of your project, you can measure the level of agreement between the model's predictions and the expert annotations. This information will give you valuable insights and help you identify any potential issues or areas for improvement.
3. Use auto-annotation tools to segment areas of interest
Annotating a tissue sample point by point is very time-consuming. Fortunately, an auto-annotation tool can be used to quickly outline regions of interest. These tools are typically based on convolutional neural networks designed to recognize patterns in images. While they won’t classify tissues or objects without some additional training, they do recognize common shapes and patterns out of the box.
For instance, a generic auto-annotation can automatically detect where one cell type ends and another begins. We can then map these regions to specific classes and add some additional attributes if necessary.

Later on, this tool can be trained to recognize and highlight specific structures, such as tissue-folds in histopathological samples. All we need is an initial training set based on “generic” auto-annotations.
4. Choose a tool that can handle large files and medical imaging formats
Some annotation tools are not able to handle large files or medical imaging formats. For example, you may experience slow loading times or other performance issues when working with large files. You may be unable to open or view certain types of medical imaging files, especially multichannel slides, Z-stack or time lapse series.
A good annotation platform should let you use video annotation features (such as frame interpolation) for time-lapse microscopy. It is also important that your training data engine supports SVS technology. By breaking a file into tiles, these viewers and platforms let you view and annotate images at different zoom levels without loss of resolution

By choosing a tool that is specifically designed to handle medical imaging formats, you can avoid many technical issues and ensure that you are able to annotate your digital pathology slides efficiently.
Still, with some tissue samples you may want to split one slide into multiple images for convenience. In V7, you can automatically crop slides and generate new datasets based on individual tiles with webhooks.
Read more: How to Use V7 Workflows to Split Large Images Into Patches
5. Leverage existing datasets that are available online
Most AI platforms require lots of data for training an accurate model. And some pathology samples may need to be pre-processed, normalized, and cleaned before the annotation process even starts. That’s why coming up with proof of concept machine learning pipelines can be so challenging.
But why start from scratch when you can build on what's already there?

If you want to test if AI can detect specific cells, bacteria, or microvascular changes it is best to try it out on existing open datasets for computer vision tasks. You can browse open-source digital pathology files to compare different annotation techniques or labels used for specific cases. With a V7 account, you can also import them, make adjustments, and train your own models. In some cases, you can create cell detection or tissue segmentation models with as little as 10 slides.
⚠️ Keep in mind that there may be a high degree of variability in the digital pathology datasets available online. If you collect your datasets from multiple sources, they often come in different shapes, sizes, and formats, which may require some advanced data wrangling techniques. Additionally, medical information is highly sensitive, and strict regulations (such as HIPAA) exist to protect patients' personal and medical information from being shared without their consent. Make sure to read the terms of use and privacy policies of any dataset you use.
Conclusion
Machine learning has been a key driving force in the development of digital pathology. The advancements in computer vision have helped to reduce the time and cost of research, diagnosis, and treatment. By combining digital microscopy and artificial intelligence, clinicians and researchers can now analyze slides and detect features of interest faster than ever.
But, AI models are only as good as the data they are trained on. So, the quality of the data is paramount.
That’s why the proper annotation of digital pathology images is becoming an increasingly important skill.
Automation tools, such as Auto-Annotate, can help you to speed up the labeling process but it is also essential to pay attention to:
Label management and annotation classes
Annotation workflow design
Your review and QA processes
By following the guidelines from this article and using the right software, you can ensure that your project is successful.
If you want to learn more, you might also be interested in: